Exercises Part 3 | Analyzing Neural Time Series Data
Exercises 10 | Convolution
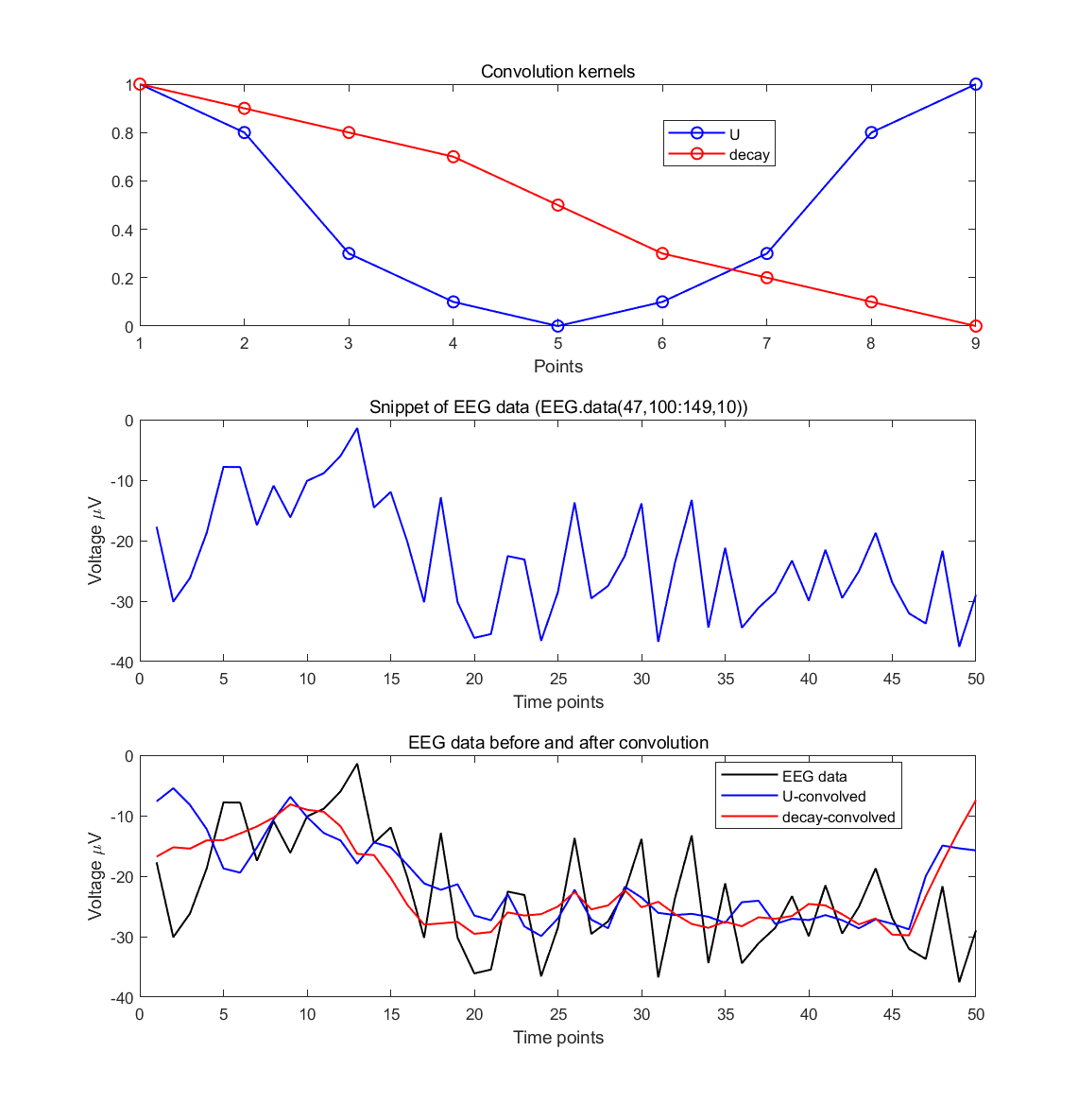
10.6.1
- Create two kernels for convolution: one that looks like a U and one that looks like a decay function. There is no need to be too sophisticated in generating, for example, a Gaussian and an exponential; numerical approximations are fine.
%% Exercises 10 |
10.6.2
- Convolve these two kernels with 50 time points of EEG data from one electrode. Make a plot showing the kernels, the EEG data, and the result of the convolution between the data and each kernel. Use time-domain convolution as explained in this chapter and as illustrated in the online Matlab code. Based on visual inspection, what is the effect of convolving the EEG data with these two kernels?
%% 2 |
Exercises 11 | Fourier Transform
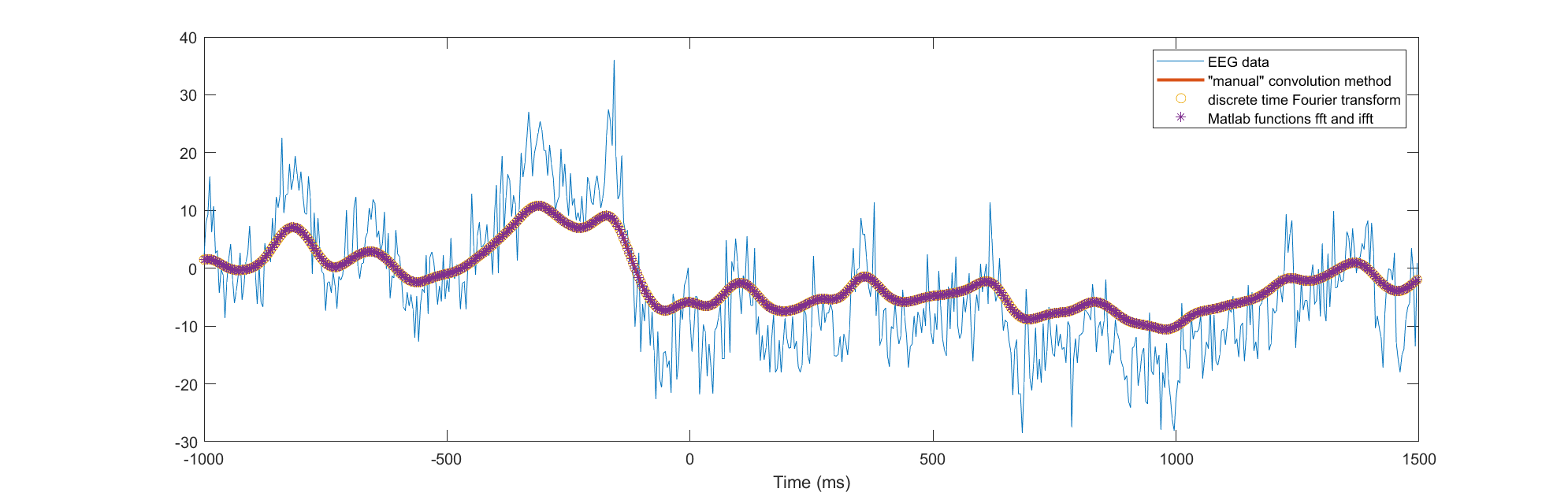
11.12.1
Reproduce the top three panels of figure 11.12 three times.
First, perform time-domain convolution using the “ manual ” convolution method shown in chapter 10.
Second, perform frequency-domain convolution using the discrete time Fourier transform presented at the beginning of this chapter.
- Finally, perform frequency-domain convolution using the Matlab functions fft and ifft (do not use the function conv). (You can optionally reproduce the bottom panel of figure 11.12 for the frequency domain analyses; keep in mind that the power scaling is for display purposes only.)
%% Exercises 11 |
11.12.2
From the three sets of Matlab code you have for reproducing figure 11.12 , run a computation time test. That is, time how long it takes Matlab to perform 1000 repetitions of each of the three methods for computing convolution that you generated in the previous exercise (do not plot the results each time). You can use the Matlab function pairs tic and toc to time a Matlab process. Plot the results in a bar plot, similar to figure 11.8 .
%% Exercises 11 |
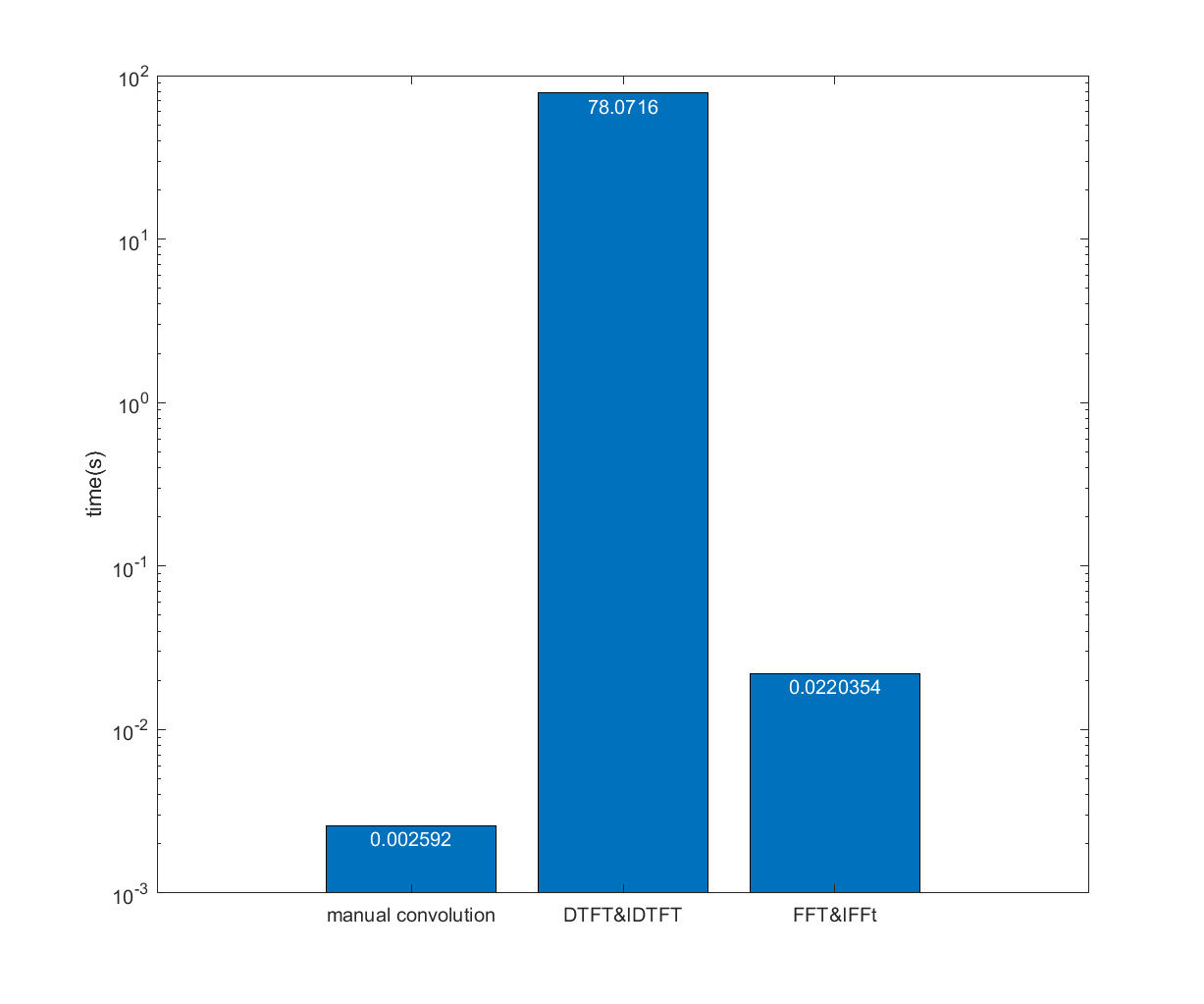
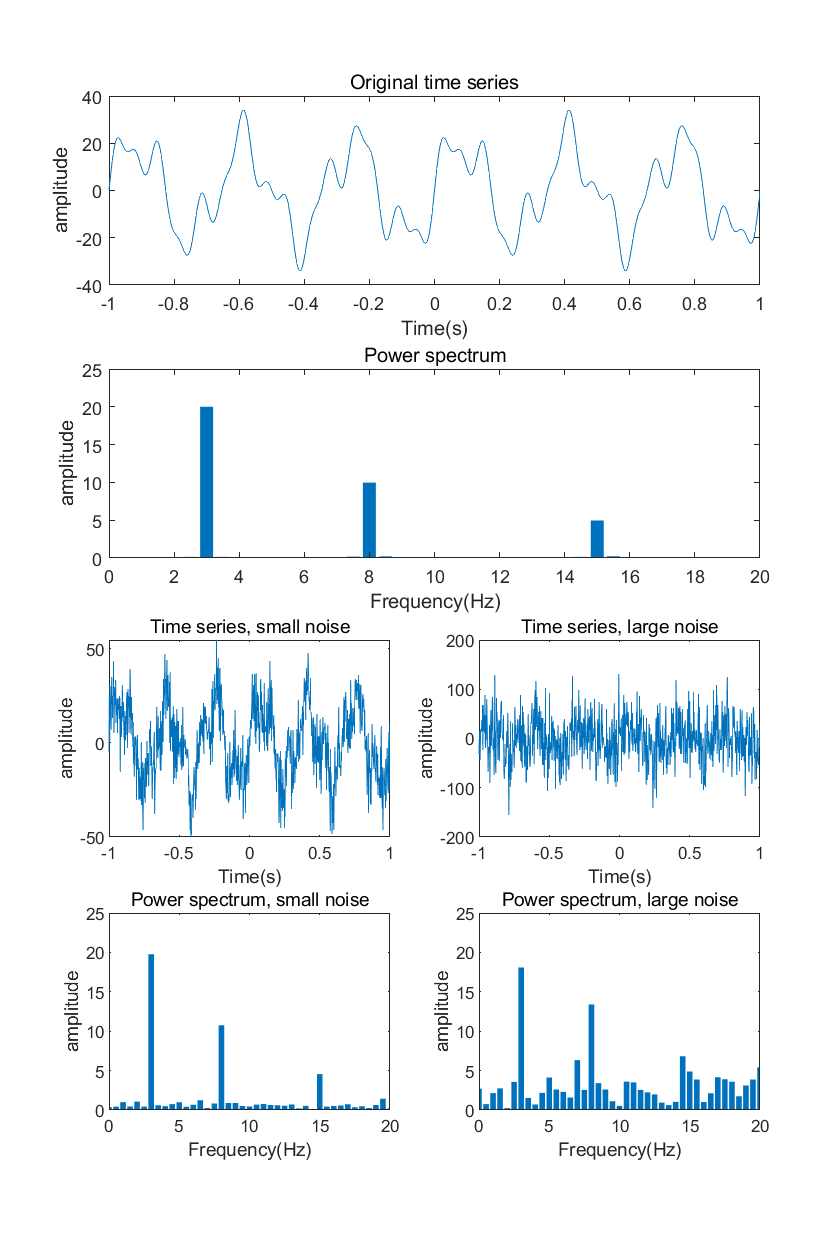
11.12.3
Generate a time series by creating and summing sine waves, as in figure 11.2B . Use between two and four sine waves, so that the individual sine waves are still somewhat visible in the sum. Perform a Fourier analysis (you can use the fft function) on the resulting time series and plot the power structure. Confirm that your code is correct by comparing the frequencies with nonzero power to the frequencies of the sine waves that you generated. Now try adding random noise to the signal before computing the Fourier transform. First, add a small amount of noise so that the sine waves are still visually recognizable. Next, add a large amount of noise so that the sine waves are no longer visually recognizable in the time domain data. Perform a Fourier analysis on the two noisy signals and plot the results. What is the effect of a small and a large amount of noise in the power spectrum? Are the sine waves with noise easier to detect in the time domain or in the frequency domain, or is it equally easy/difficult to detect a sine wave in the presence of noise?
%% Exercises 11 |
Exercises 12 | Morlet wavelets
Create a family of Morlet wavelets ranging in frequency from 2 Hz to 30 Hz in five steps.
Select one electrode from the scalp EEG dataset and convolve each wavelet with EEG data from all trials from that electrode. Apply the Matlab function real to the convolution result, as in
convol_result=real(convol_result). This will return the EEG data bandpass filtered at the peak frequency of the wavelet. You learn more about why this is in the next chapter.Average the result of convolution over all trials and plot an ERP corresponding to each wavelet frequency. Each frequency should be in its own subplot.
Plot the broadband ERP (without any convolution). Thus, you will have six subplots in one figure. How do the wavelet-convolved ERPs compare with the broadband ERP? Are there dynamics revealed by the wavelet-convolved ERPs that are not apparent in the broadband ERP, and are there dynamics in the broadband ERP that are not apparent in the waveletconvolved ERPs? Base your answer on qualitative visual inspection of the results; statistics or other quantitative comparisons are not necessary.
%% Exercise 12 |
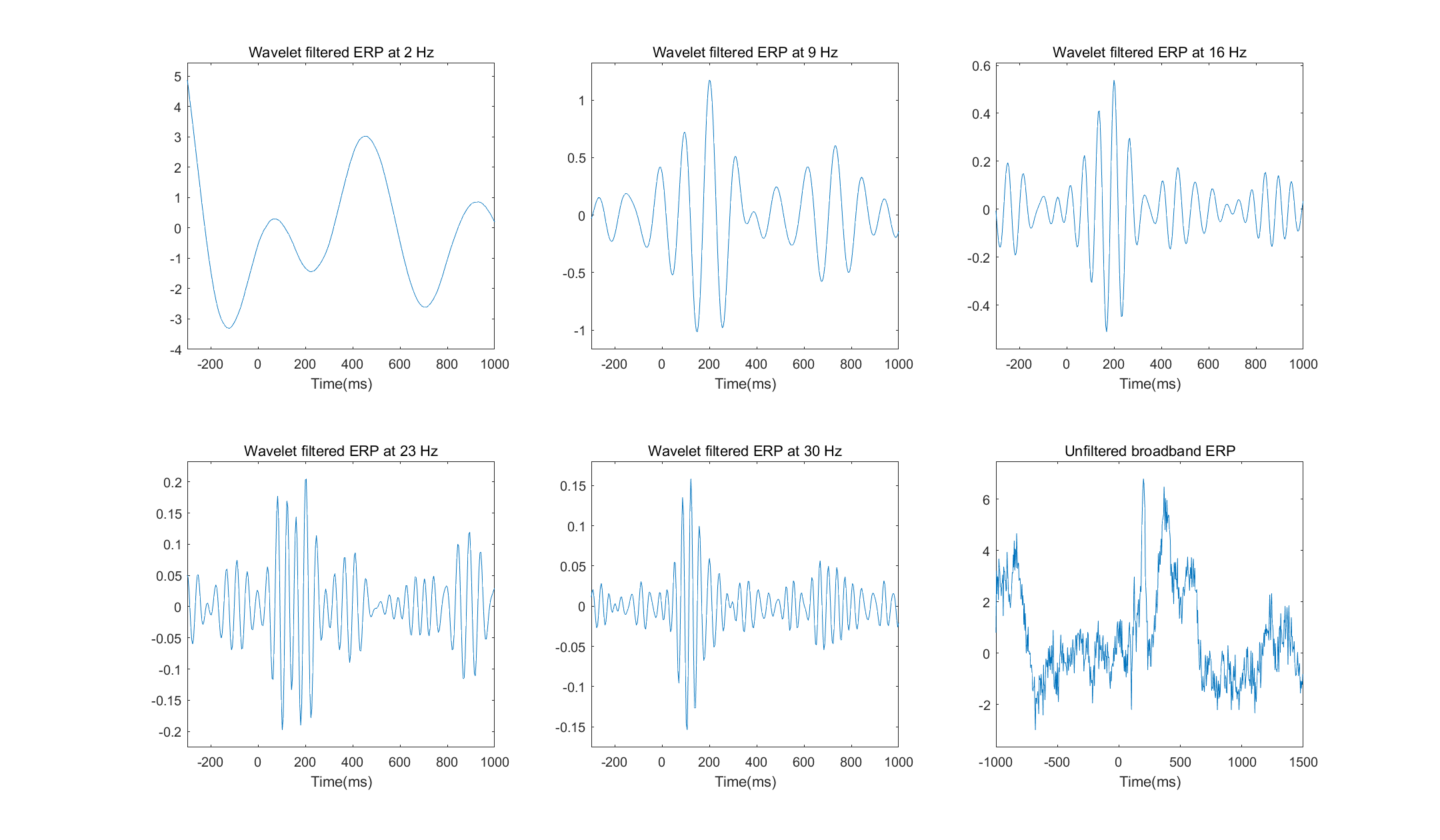
Exercises 13 | complex Morlet wavelets
1. Create a family of complex Morlet wavelets
Create a family of complex Morlet wavelets ranging in frequencies from 2 Hz to 30 Hz in five steps.
%% Exercises 13 |
2. Convolve each wavelet with EEG data
Convolve each wavelet with EEG data from all electrodes and from one trial.
3. Extract power and phase
Extract power and phase from the result of complex wavelet convolution and store in a time × frequency × electrodes × power/phase matrix (thus, a 640 × 5 × 64 × 2 matrix).
%% 2. Convolve each wavelet with EEG data from all electrode and from one trial |
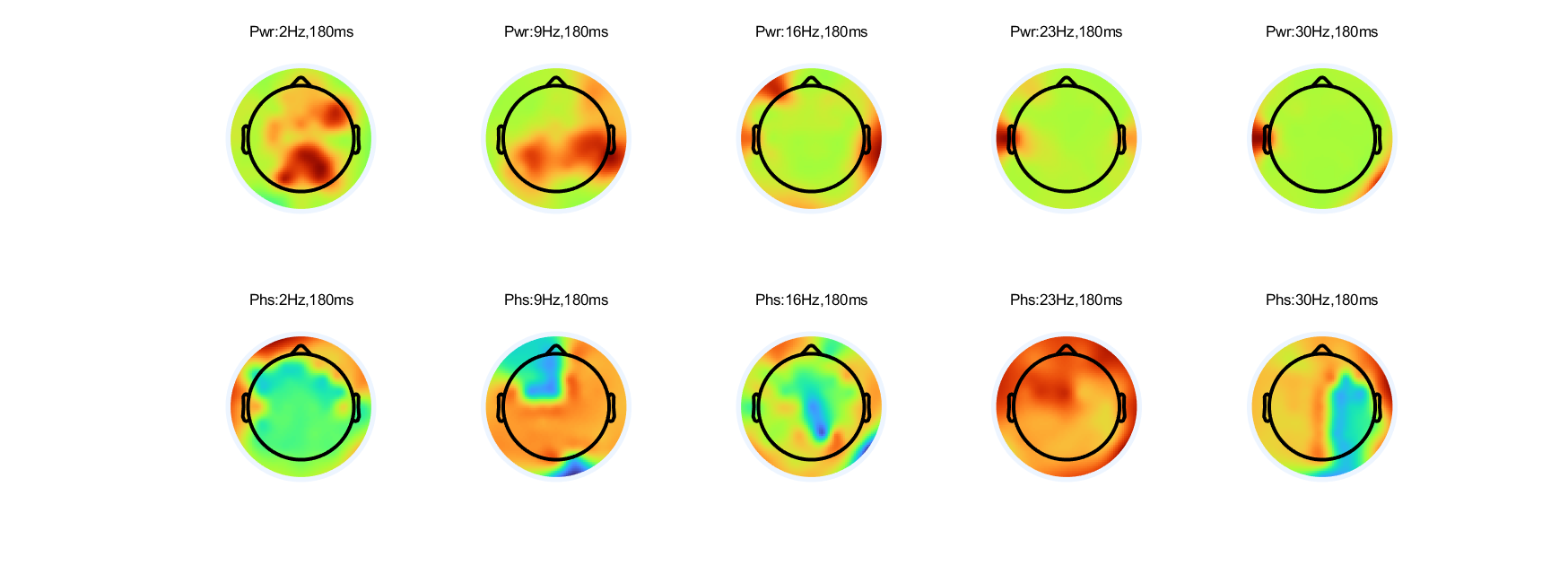
4. Make topographical plots of power and phase
Make topographical plots of power and phase at 180 ms at all frequencies (hint: you may need to use the squeeze function to remove singleton dimensions). Arrange the plots in one figure with five columns for frequency and two rows for power/phase. Put labels in the plot so it is clear which topographical maps correspond to which frequencies.
%% make topographical features in power ans phase at 180 ms |
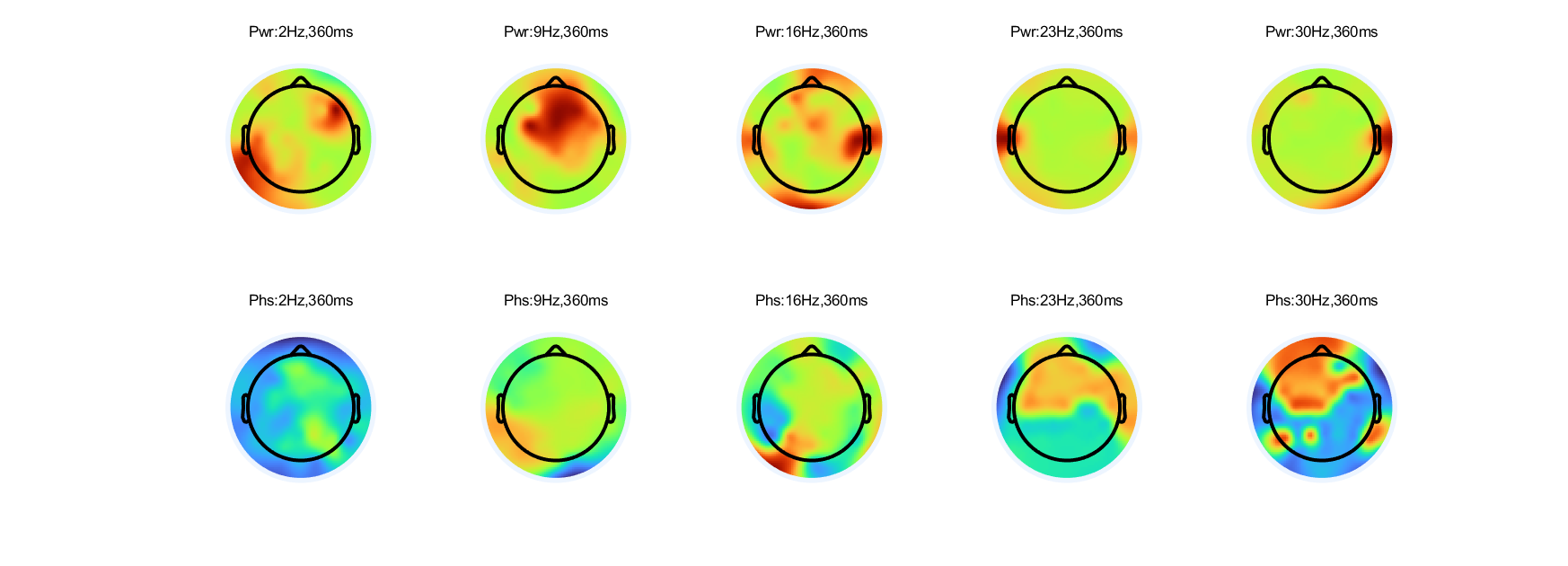
5. Repeat step 4 for activity at 360 ms.
%% make topographical features in power ans phase at 360 ms |
6. Think
Are there any prominent topographical features in power or in phase? - Yes.
Do these differ for different frequencies? - Yes.
Do power and phase have similar topographical distributions? - It doesn’t look very similar.
Is there any reason to suspect that they might have similar or different topographies?
(From ChatGPT for reference, GPT is not provided with corresponding pictures)
When analyzing EEG data, particularly through time-frequency decomposition, you might observe various topographical features in both power and phase distributions across the scalp. These features can provide insights into underlying neural processes. Let’s break down each question:
(1) Are there any prominent topographical features in power or in phase?
- Power Topography: Power topography represents the distribution of signal power across the scalp. Prominent features in power maps could indicate areas of strong neural activity, which might correspond to specific cognitive or sensory processes. For example, increased power in the occipital regions might be related to visual processing.
- Phase Topography: Phase topography shows the distribution of phase angles across the scalp. Prominent features in phase topography could indicate phase synchronization between regions, which might be associated with communication between different brain areas.
(2) Do these differ for different frequencies?
- Yes, topographical features often differ for different frequencies. Different frequencies in EEG are associated with different types of neural activity (e.g., alpha waves in the 8-12 Hz range are often related to relaxation and closed eyes, while beta waves in the 13-30 Hz range are associated with active thinking or focus). The power and phase topographies can change significantly across different frequency bands, reflecting the different functional roles of these frequencies.
(3) Do power and phase have similar topographical distributions?
- Power and phase distributions are often not identical. Power topography might highlight regions with strong amplitude, while phase topography might emphasize regions where the timing of oscillatory activity is synchronized across electrodes. However, there can be similarities, especially in regions where strong power is accompanied by consistent phase alignment (e.g., in tasks requiring coordinated neural activity across brain regions).
(4) Is there any reason to suspect that they might have similar or different topographies?
- Similar Topographies: Power and phase could have similar topographies in cases where high power is associated with phase-locked activity across trials or regions. For instance, during sensory processing, a stimulus might evoke both strong power and phase synchronization in relevant brain areas.
- Different Topographies: Conversely, power and phase might differ in their topographical distributions due to different underlying mechanisms. High power might occur in one region without strong phase alignment, especially if the neural activity is more localized or independent across trials. Additionally, phase synchronization can occur without strong power, especially in low-amplitude oscillations that are critical for certain cognitive processes.
Summary:
- Power and phase topographies can both reveal important aspects of neural activity, but they often provide different insights. Power maps tend to show where neural activity is strong, while phase maps can indicate where different regions are temporally synchronized. The topographies of power and phase may differ depending on the frequency of interest and the specific neural processes involved. Understanding these distributions and their relationships is key to interpreting EEG data in the context of neural dynamics.
7. Create a circular colormap
Because phase values are circular ( – π and + π are identical), most color maps are inappropriate because they suggest that – π and + π are very different values (represented, e.g., by blue and red colors). Create a circular colormap that can be used for phase values. You can do this by setting the red, green, and/or blue values to be a cosine function rather than a linear function. Recreate the phase topographical maps. Do they look any different with the new color maps?
%% 自定义用于相位的环形colormap,使得-pi和pi对应的颜色相同 |
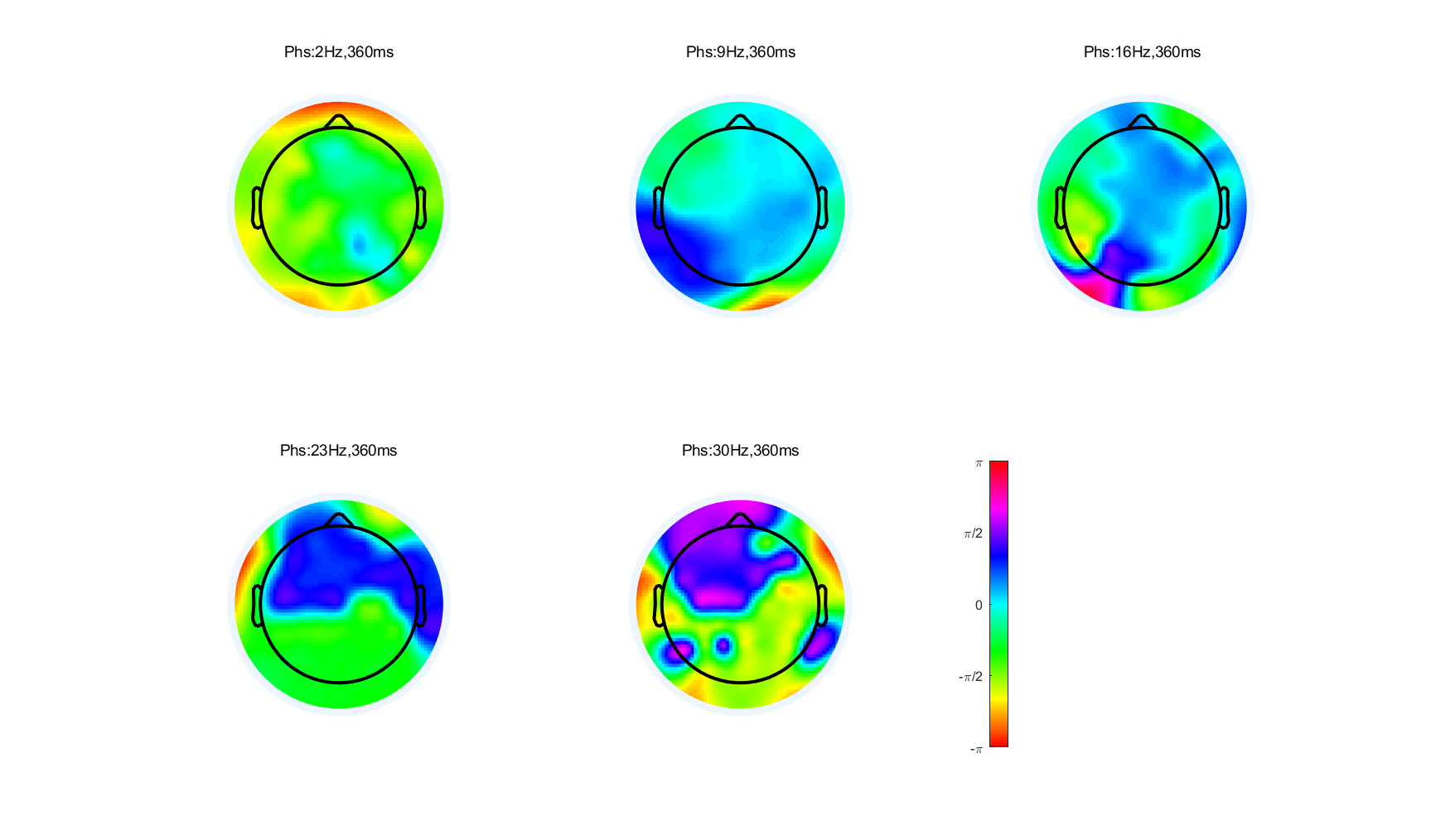
Exercises 14 | filter-Hilbert
- Pick two frequencies (e.g., 5 Hz and 25 Hz) and one electrode and perform complex Morlet wavelet convolution and filter-Hilbert using those two frequencies as the peak/center frequencies for all trials. Plot the resulting power and the bandpass-filtered signal (that is, the real component of the analytic signal) from each method. Plot one single trial (you can choose the trial randomly but plot the same trial for both methods) and then plot the average of all trials. Describe some similarities and differences between the results of the two time-frequency decomposition methods.
%% Exercises 14 |
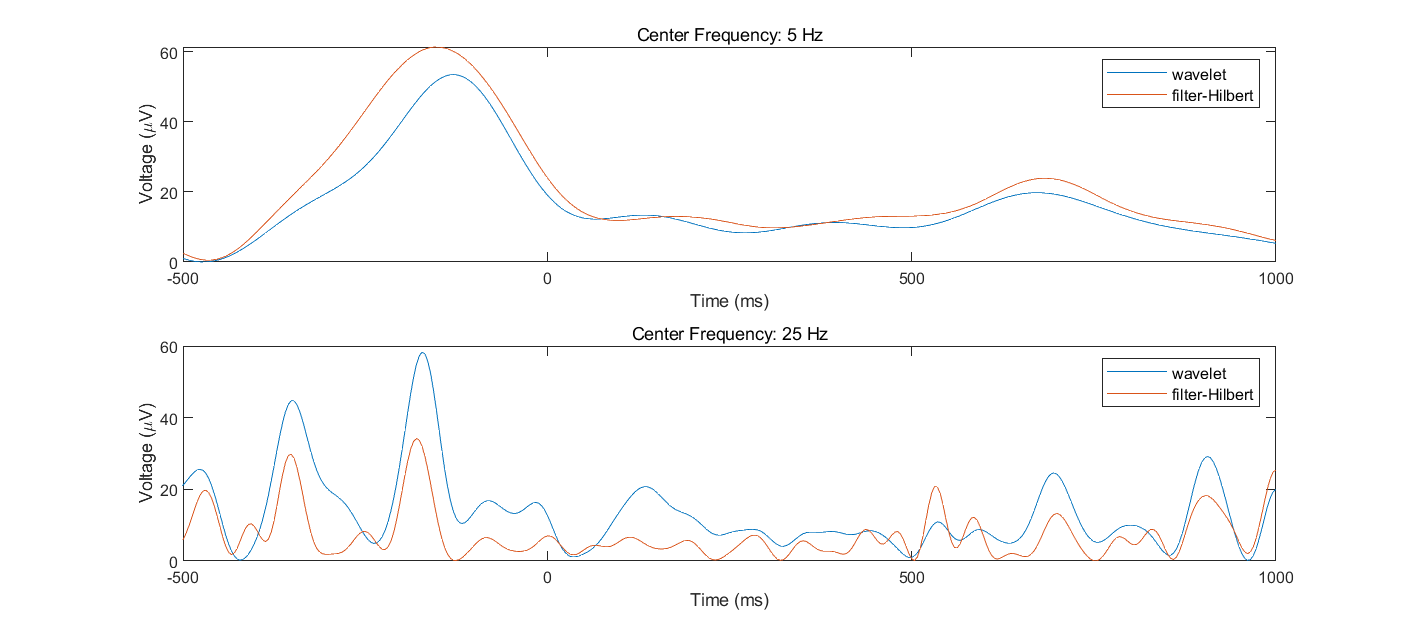
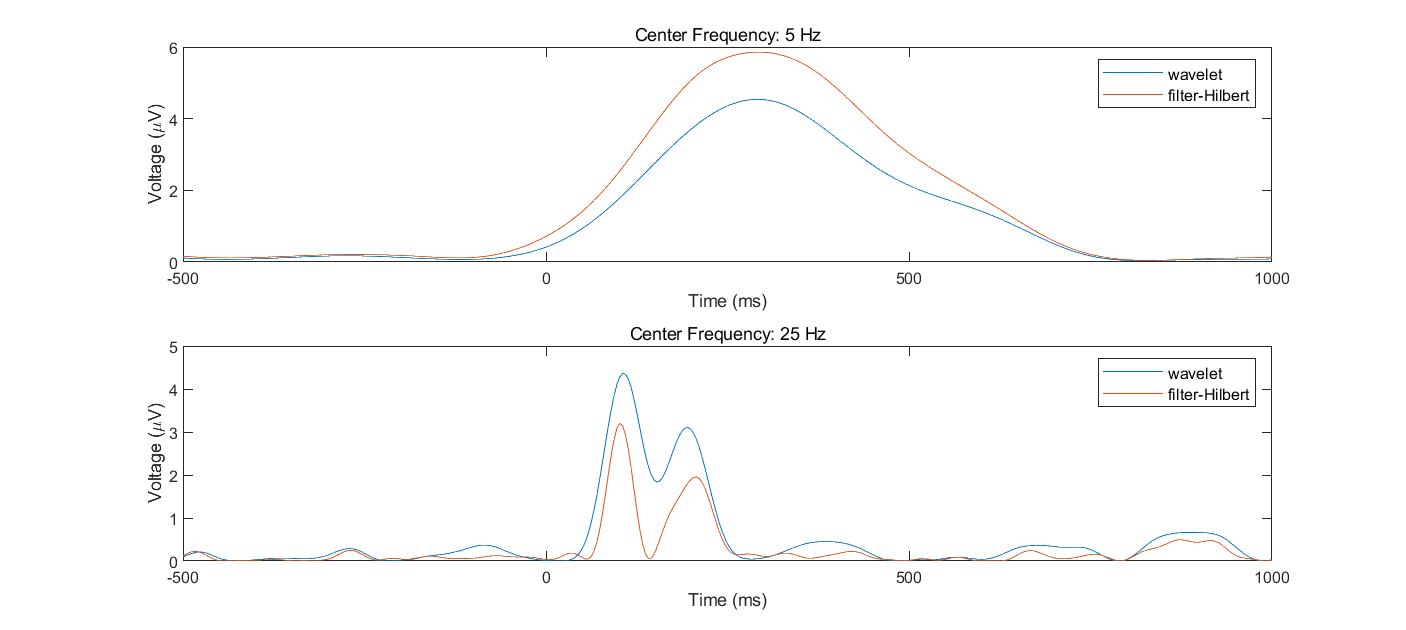
- Modify the wavelet and filter settings (but keep the peak/center frequencies the same) until these two methods produce very similar results. Next, modify the wavelet and filter settings (except the peak/center frequencies) to make the results different (stay within a reasonable range of parameter settings; they do not need to look dramatically different). Which parameters did you change to make the results look more similar versus more different? How different are the results, and would you consider this a meaningful difference? What does this difference tell you about when to use specific parameter settings for wavelet convolution and the filter-Hilbert method?
尝试改变的参数:
- wavelet:
- numcycles:wavelet包含的周期数目参数。包含的周期数目越多,wavelet频带越窄,频率分辨率越高
- filter-Hilbert:
- freqspread:带宽参数。带宽越大,频率分辨率越低
- transwid:过渡区宽度参数。
firls()的第一个参数,the order of the filter:决定filter kernel频率响应的精度。
Exercises 15 | The Short-Time FFT
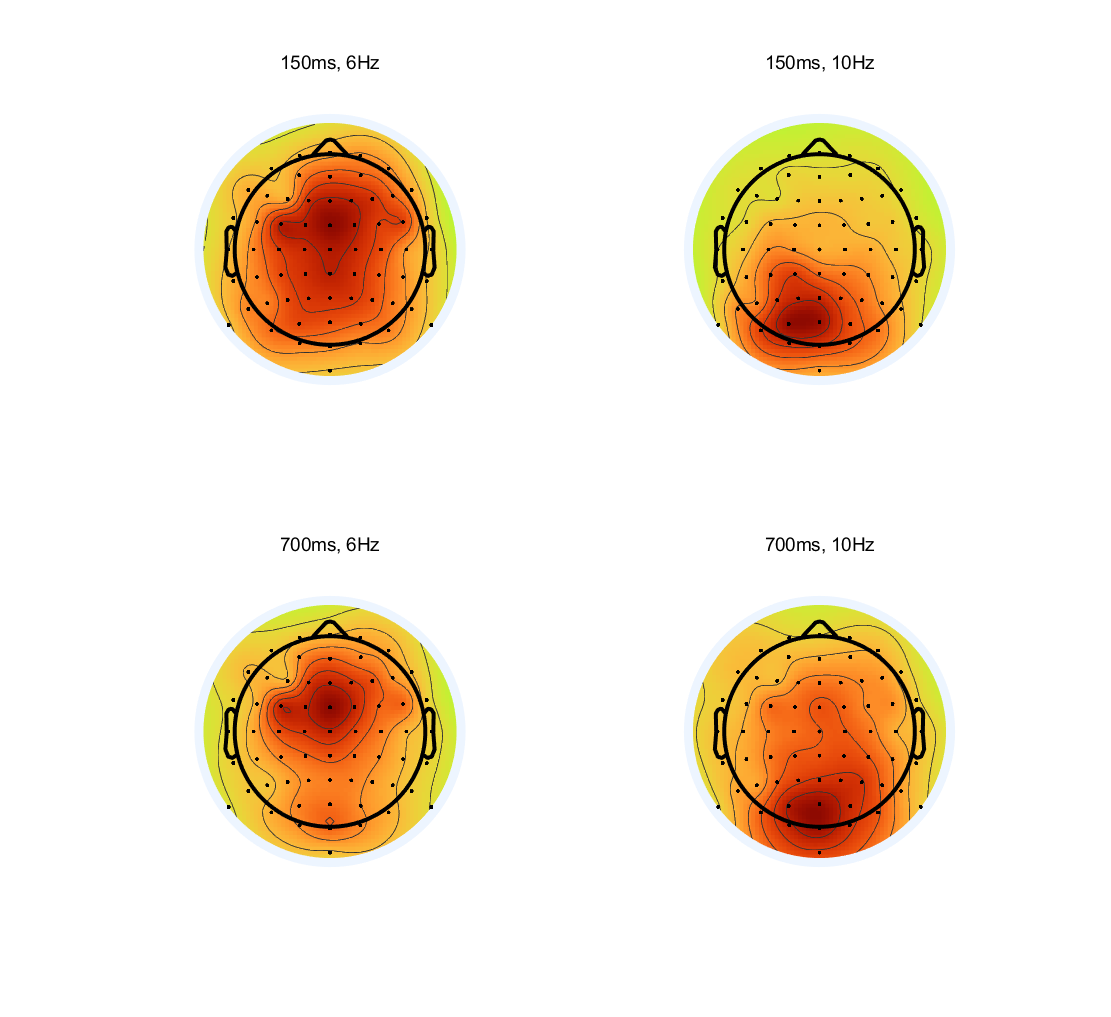
- Compute the short-time FFT at each electrode and make topographical maps of theta band (around 6 Hz) power and alpha-band (around 10 Hz) power at 150 ms and 700 ms.
%% Compute the short-time FFT at each electrode |
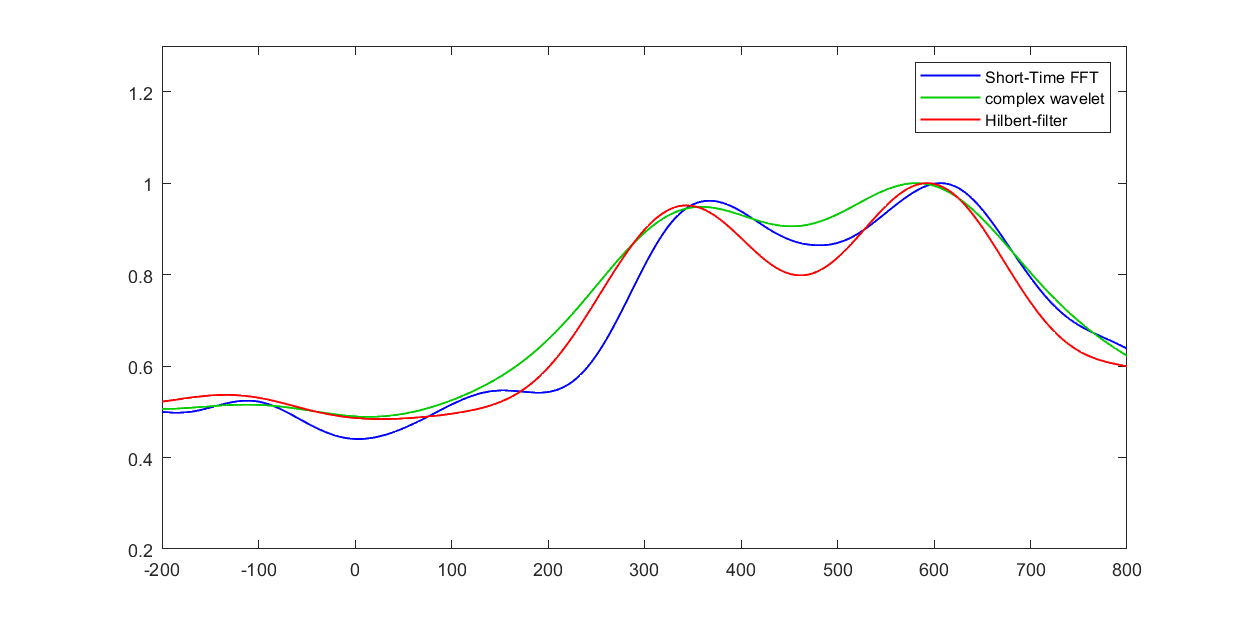
- Select one electrode and one frequency and compute power over time at that electrode and that frequency using complex wavelet convolution, filter-Hilbert, and the short-time FFT. Plot the results of these three time-frequency decomposition methods in different subplots of one figure. Note that the scaling might be different because no baseline normalization has been applied. How visually similar are the results from these three methods? If the results from the three methods are different, how are they different, and what parameters do you think you could change in the three methods to make the results look more or less similar?
%% Select one electrode and one frequency and compute power over time at that electrode and that frequency |
Exercises 16 | Multitaper
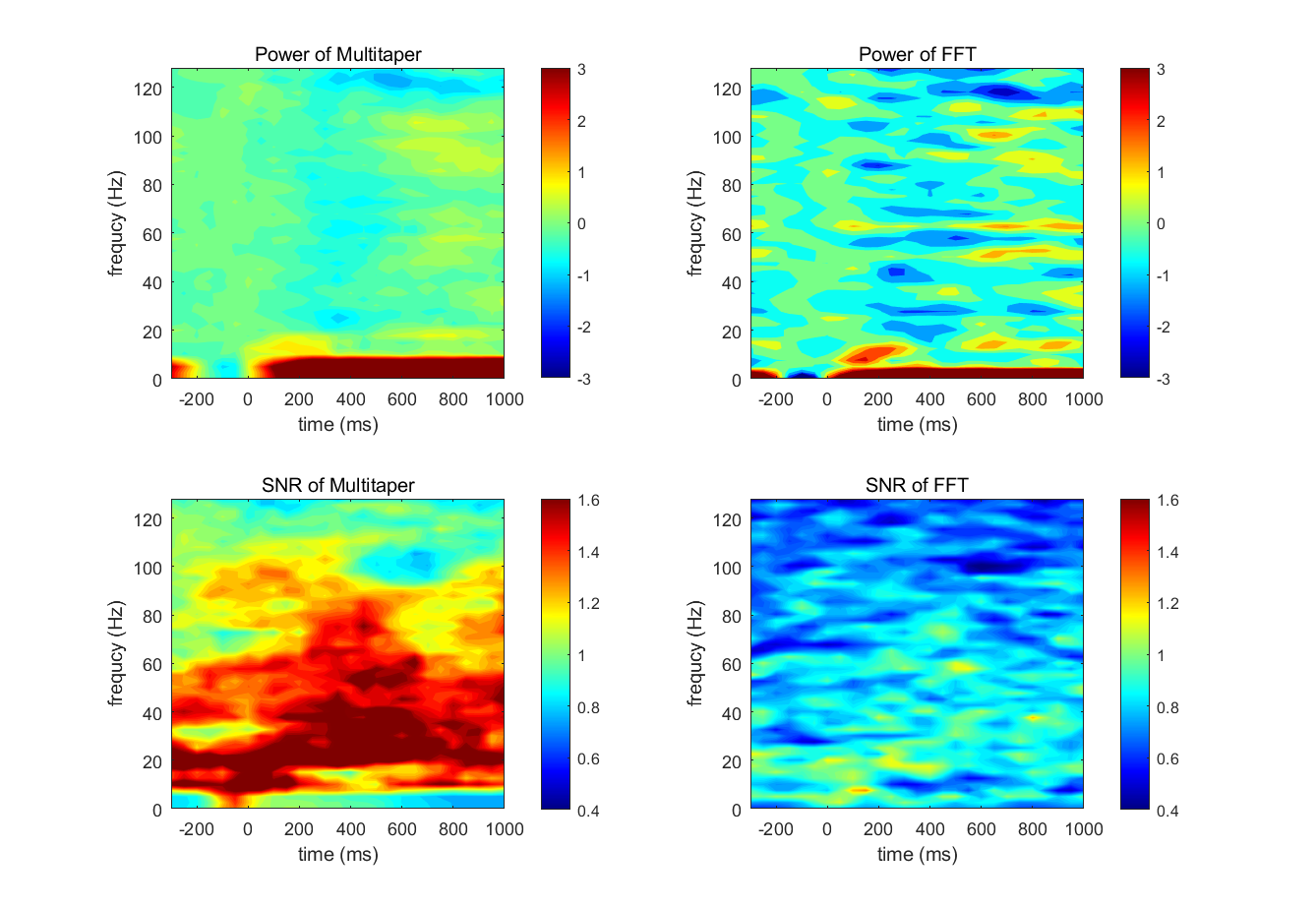
Pick one electrode and compute a time-frequency map of power using both the multitaper method and the short-time FFT. Store all of the power values for all of the trials.
Next, compute a time-frequency map of signal-to-noise ratio. The signal-to-noise ratio of power is discussed more in chapter 18, but it can be estimated as the average power at each time-frequency point across trials, divided by the standard deviation of power at each time-frequency point across trials.
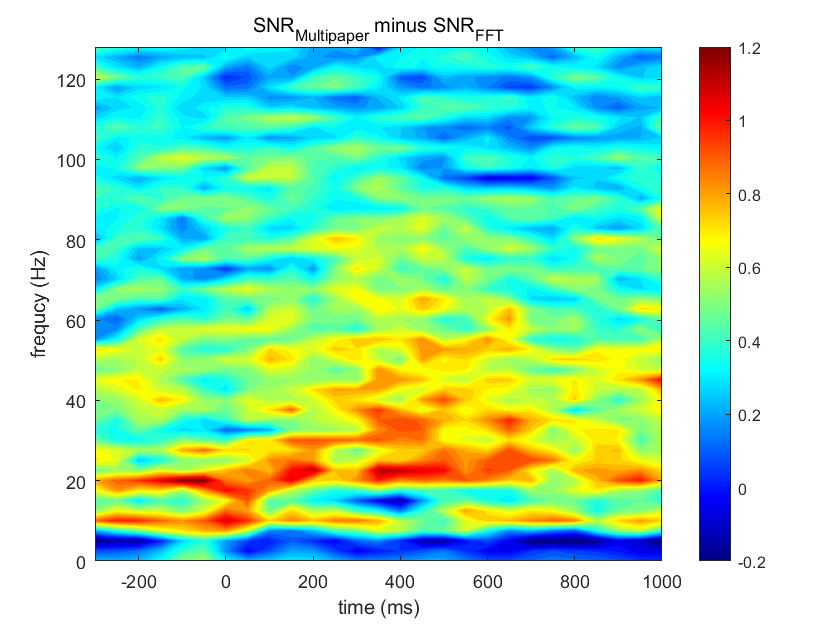
- Make time-frequency plots of power and signal-to-noise ratio from the two methods. Make another plot in which you subtract the signal-to-noise plots between the two methods. Are there any noticeable differences between the signal-to-noise results when the multitaper method versus the short-time FFT is used?
%% Pick one electrode and compute a time-frequency map of power using both the multitaper method and the short-time FFT. |
- Select two frequencies, one relatively low and one relatively high (e.g., 8 Hz and 60 Hz), and compare the power time series and signal-to-noise time series in these frequency bands from the two methods in a separate figure, using line plots. Comment on the differences if there are any.
%% Select two frequencies, one relatively low and one relatively high (e.g., 8 Hz and 60 Hz), |
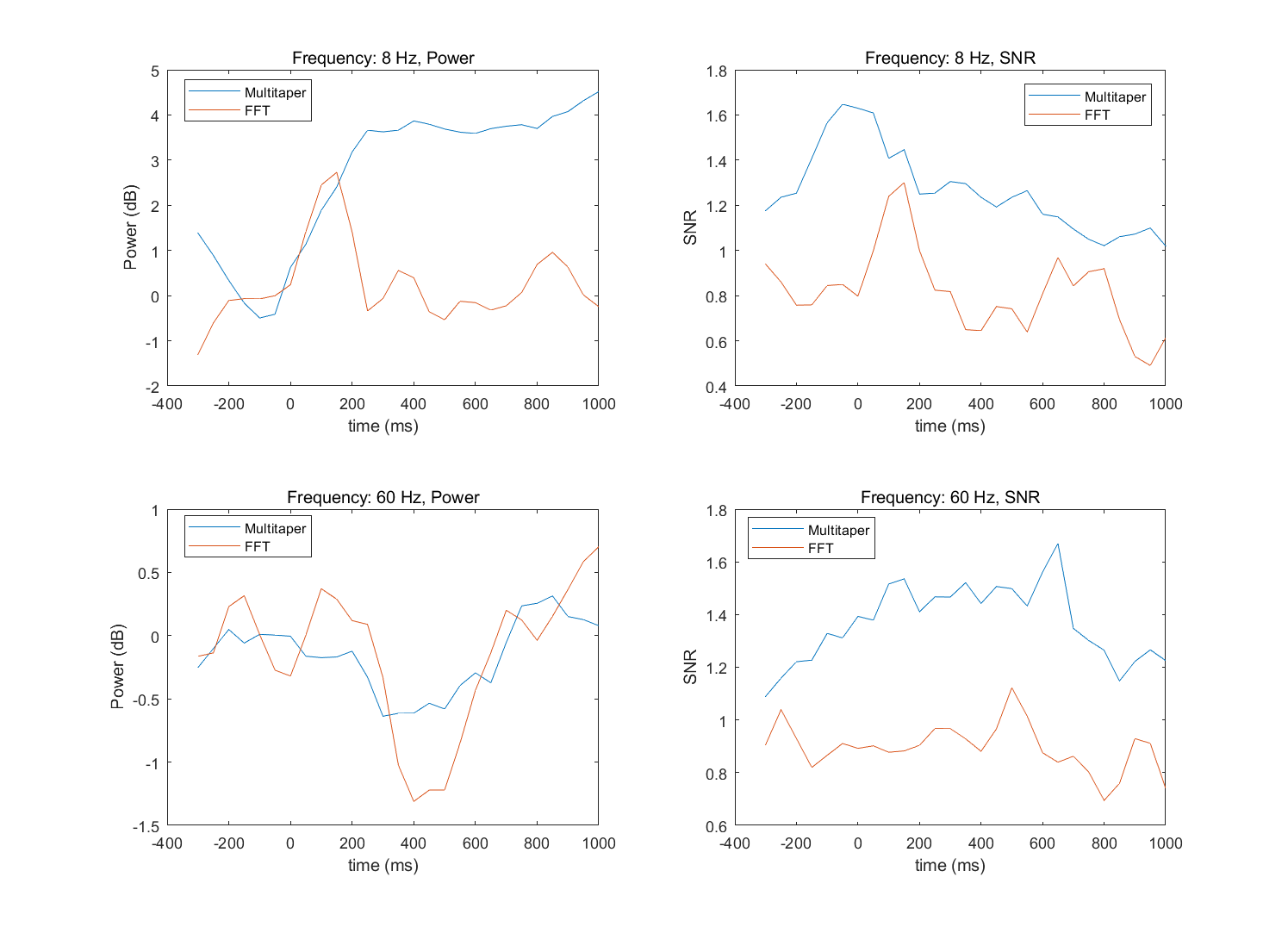
- Multitaper常用于低信噪比的情况,如高频活动或功率的单试次估计。通过使用多个taper,multitaper方法在频率轴上引入了一定的平滑效应。这种平滑可以减少频率分辨率的精细度,使得频谱变得更加连续和平滑。
Exercises 18 | Baseline Normalization
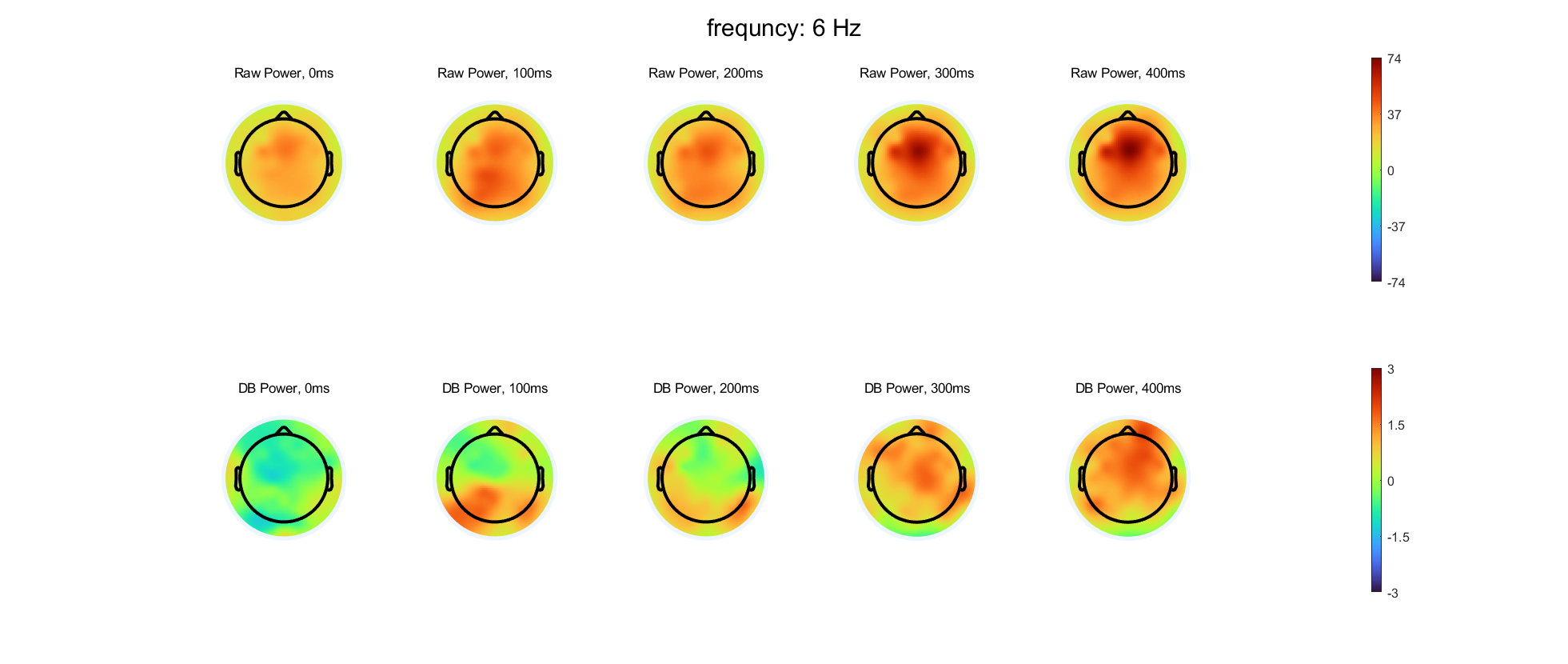
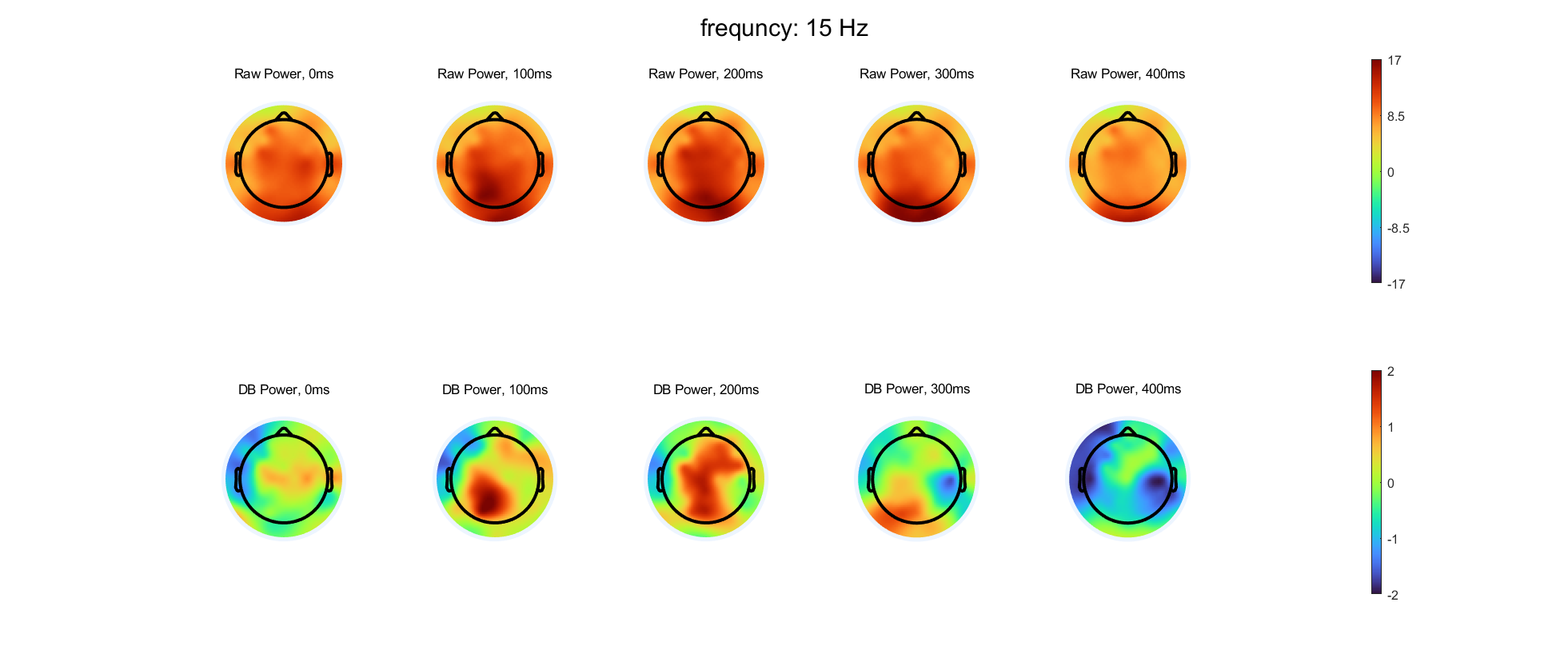
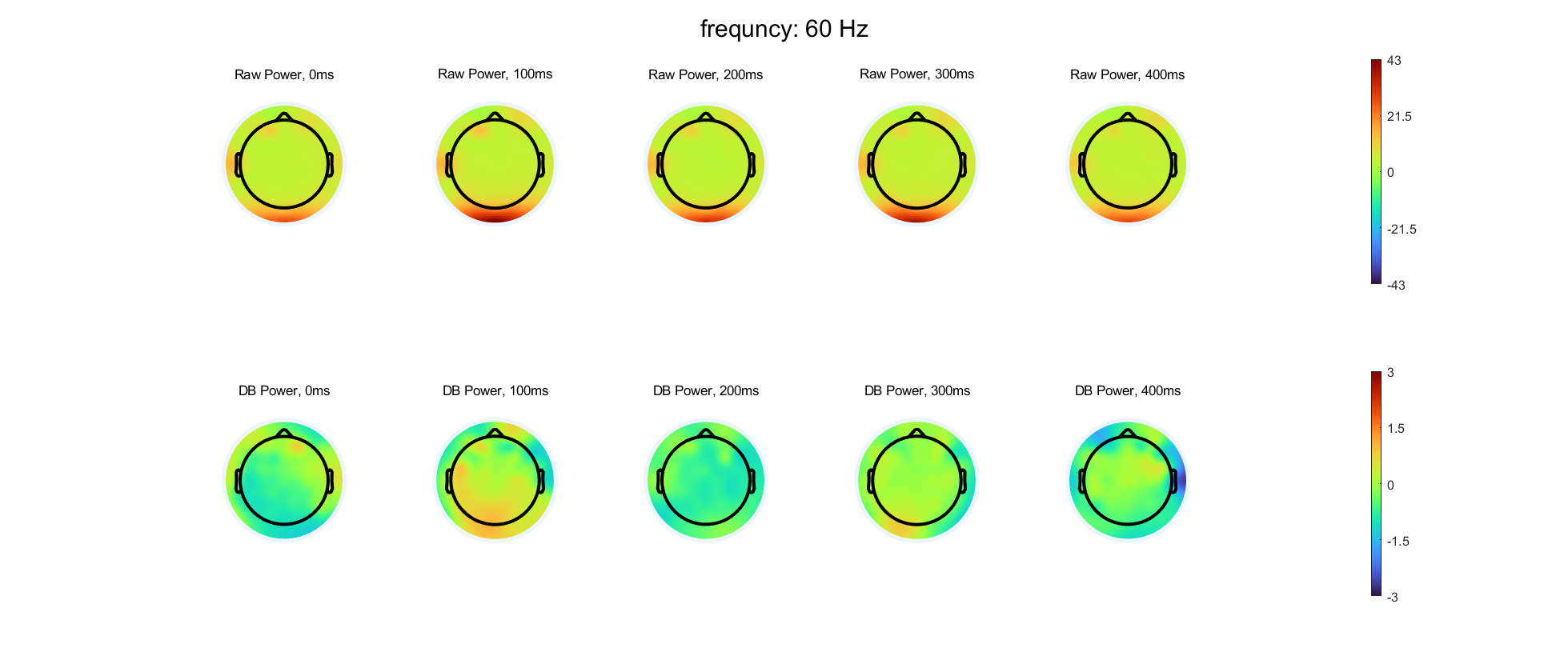
Select three frequency bands and compute time-varying power at each electrode in these three bands, using either complex wavelet convolution or filter-Hilbert. Compute and store both the baseline-corrected power and the raw non-baseline-corrected power. You can choose which time period and baseline normalization method to use.
Select five time points and create topographical maps of power with and without baseline normalization at each selected time-frequency point. You should have time in columns and with/without baseline normalization in rows. Use separate figures for each frequency. The color scaling should be the same for all plots over time within a frequency, but the color scaling should be different for with versus without baseline normalization and should also be different for each frequency.
- Are there qualitative differences in the topographical distributions of power with compared to without baseline normalization? Are the differences more prominent in some frequency bands or at some time points? What might be causing these differences?
%% Select three frequency bands and compute time-varying power at each electrode in these three bands, using either complex wavelet convolution or filter-Hilbert. |
Exercises 19 | ITPC (Intertrial Phase Clustering)
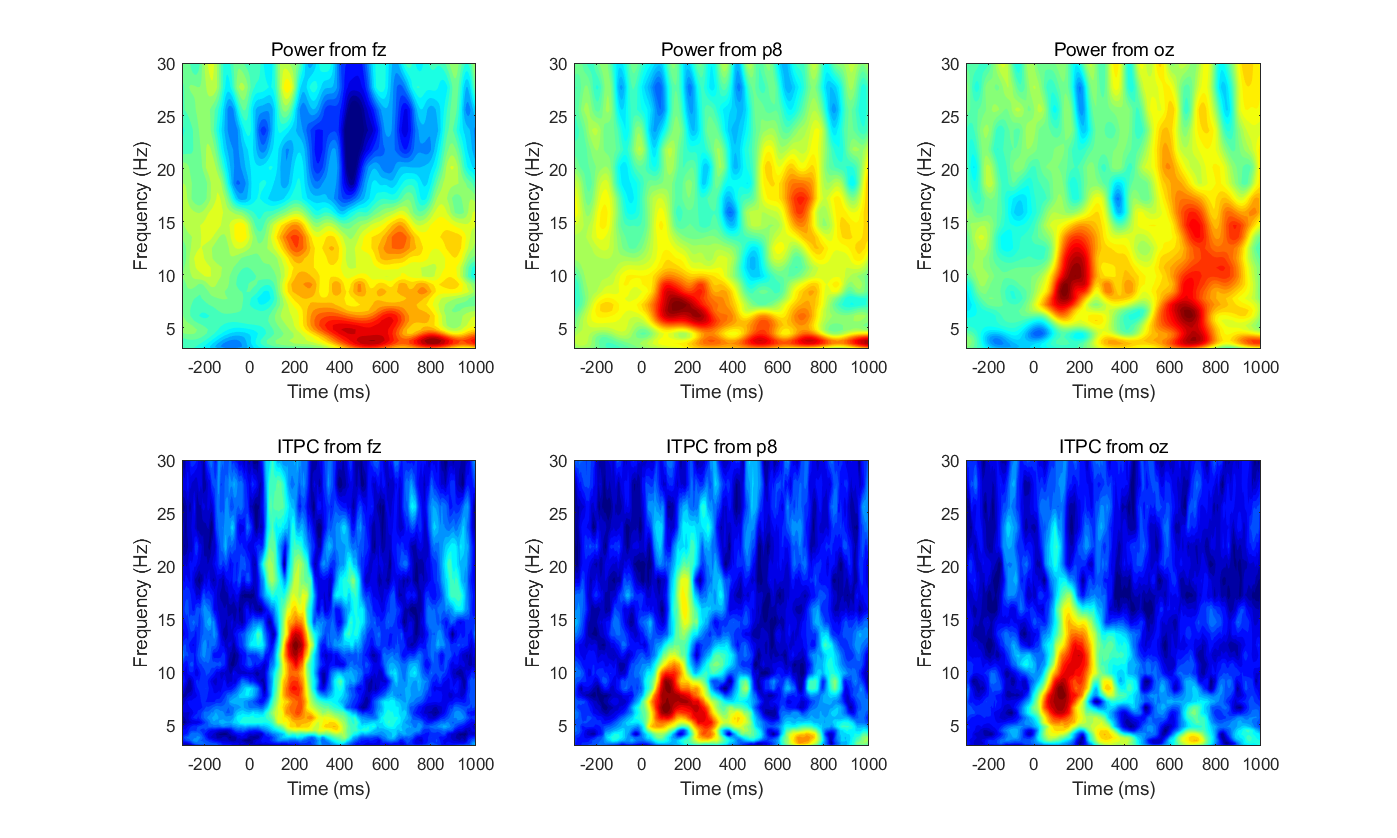
- Pick three electrodes. Compute time-frequency plots of ITPC and decibel-corrected power for these electrodes, using either complex Morlet wavelet convolution or the filter-Hilbert method. Plot the results side by side for each electrode (power and ITPC in subplots; one figure for each electrode). Are the patterns of results from ITPC and power generally similar or generally different? Do the results look more similar at some electrodes and less similar at other electrodes?
%% Pick three electrodes. Compute time-frequency plots of ITPC and decibel-corrected power for these electrodes, using either complex Morlet wavelet convolution or the filter-Hilbert method. |
- For each of these three electrodes, compute wITPCz using reaction time as the trial-varying modulator. Perform this analysis for all time-frequency points to generate time-frequency maps of the relationship between phase and reaction time. Do the time-frequency maps of wITPCz look different from the time-frequency maps of ITPC? Do you see any striking patterns in the ITPCz results, and do the results differ across the different electrodes (don ’ t worry about statistics, base your judgment on qualitative patterns)? How would you interpret the results if they were statistically significant?
Exercises 20 | Total, Phase-Locked, and Non-Phase-Locked Power and Intertrial Phase Consistency
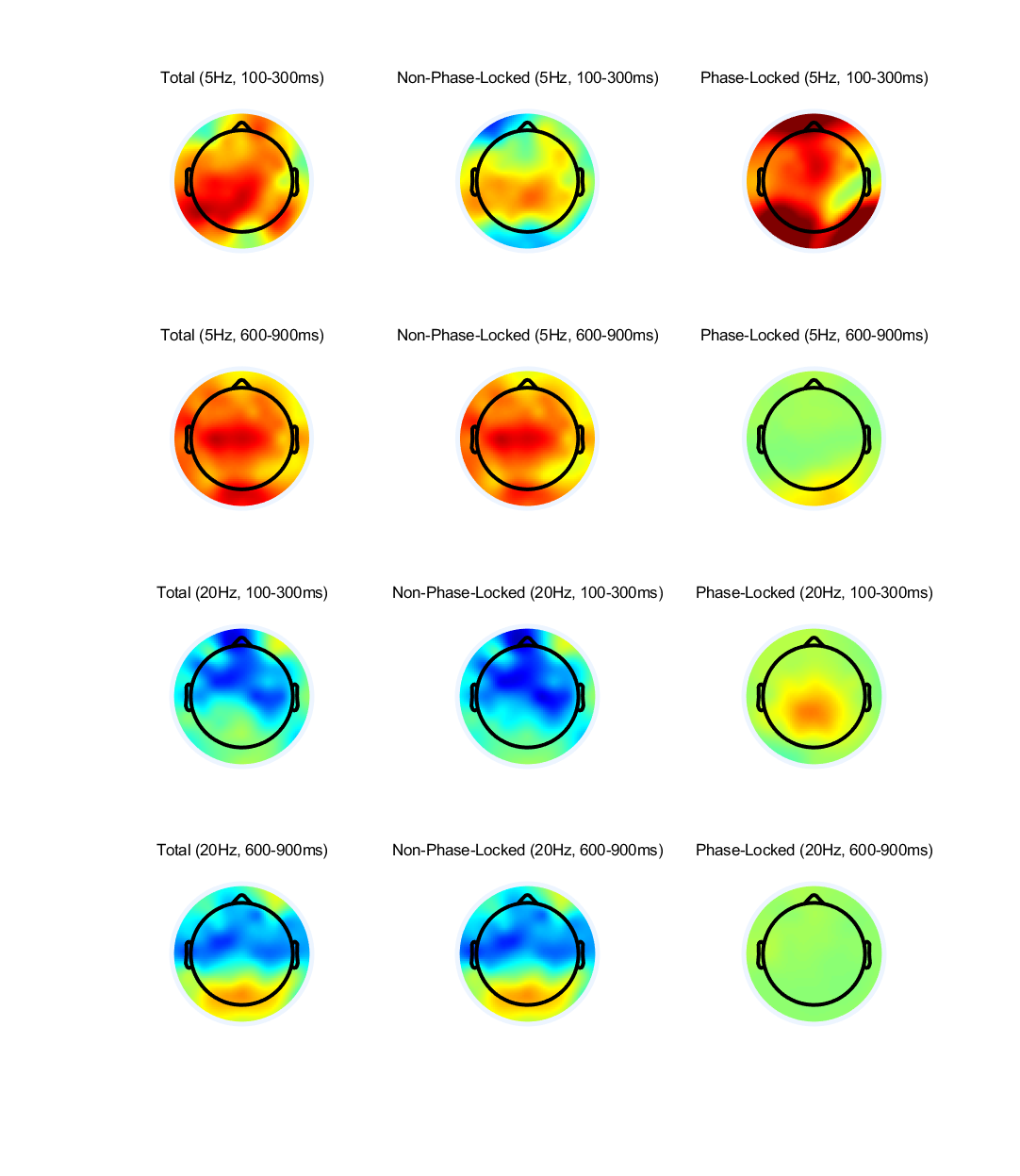
Pick two frequencies and compute total and non-phase-locked power from each electrode over time at these two frequencies. Pick two time windows, one early and one late, of several hundreds of milliseconds each (e.g., 100 – 300 ms and 500 – 800 ms) and show topographical maps of total power, non-phase-locked power, and phase-locked power from the average of all time points within these windows. Are there striking topographical differences among these results? If so, are the differences bigger or smaller in the early or the late time window? Why might this be the case?
%% Pick two frequencies and compute total and non-phase-locked power from each electrode over time at these two frequencies. |
Tips
把以ms为单位的时间转化为时间的索引index可以用
dsearchn函数:% convert requested times to indices
times2saveidx = dsearchn(EEG.times',times2save');

